| dyads_test_vs_ctrl_m1_shift0 (dyads_test_vs_ctrl_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m1; m=0 (reference); ncol1=15; shift=0; ncol=15; ahaAAAGCCCAAawh
; Alignment reference
a 61 47 97 112 130 137 15 2 8 2 148 127 98 42 47
c 28 41 31 19 10 1 6 152 148 154 6 8 26 30 44
g 33 28 10 13 6 22 136 6 1 1 0 7 10 32 27
t 40 46 24 18 16 2 5 2 5 5 8 20 28 58 44
|
| DOF5.3_MA1071.1_JASPAR_shift0 (DOF5.3:MA1071.1:JASPAR) |
 |
0.901 |
0.421 |
1.178 |
0.941 |
0.975 |
1 |
1 |
3 |
1 |
1 |
1.400 |
1 |
; dyads_test_vs_ctrl_m1 versus DOF5.3_MA1071.1_JASPAR (DOF5.3:MA1071.1:JASPAR); m=1/4; ncol2=7; w=7; offset=0; strand=D; shift=0; score= 1.4; gawAAmG--------
; cor=0.901; Ncor=0.421; logoDP=1.178; NsEucl=0.941; NSW=0.975; rcor=1; rNcor=1; rlogoDP=3; rNsEucl=1; rNSW=1; rank_mean=1.400; match_rank=1
a 247 327 396 842 850 611 66 0 0 0 0 0 0 0 0
c 247 213 128 61 79 329 52 0 0 0 0 0 0 0 0
g 262 228 146 17 0 14 767 0 0 0 0 0 0 0 0
t 244 233 331 81 70 47 116 0 0 0 0 0 0 0 0
|
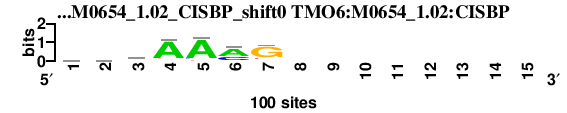
| TMO6_M0654_1.02_CISBP_shift0 (TMO6:M0654_1.02:CISBP) |
 |
0.901 |
0.421 |
1.169 |
0.941 |
0.975 |
2 |
2 |
4 |
2 |
2 |
2.400 |
2 |
; dyads_test_vs_ctrl_m1 versus TMO6_M0654_1.02_CISBP (TMO6:M0654_1.02:CISBP); m=2/4; ncol2=7; w=7; offset=0; strand=D; shift=0; score= 2.4; vawAAmG--------
; cor=0.901; Ncor=0.421; logoDP=1.169; NsEucl=0.941; NSW=0.975; rcor=2; rNcor=2; rlogoDP=4; rNsEucl=2; rNSW=2; rank_mean=2.400; match_rank=2
a 25 33 40 84 85 61 7 0 0 0 0 0 0 0 0
c 25 21 13 6 8 33 5 0 0 0 0 0 0 0 0
g 26 23 14 2 0 1 76 0 0 0 0 0 0 0 0
t 24 23 33 8 7 5 12 0 0 0 0 0 0 0 0
|
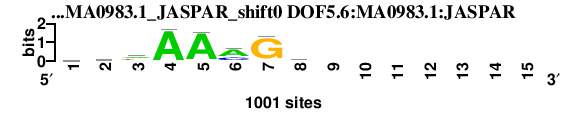
| DOF5.6_MA0983.1_JASPAR_shift0 (DOF5.6:MA0983.1:JASPAR) |
 |
0.769 |
0.410 |
1.682 |
0.908 |
0.933 |
3 |
3 |
2 |
3 |
3 |
2.800 |
3 |
; dyads_test_vs_ctrl_m1 versus DOF5.6_MA0983.1_JASPAR (DOF5.6:MA0983.1:JASPAR); m=3/4; ncol2=8; w=8; offset=0; strand=D; shift=0; score= 2.8; vawAAmGt-------
; cor=0.769; Ncor=0.410; logoDP=1.682; NsEucl=0.908; NSW=0.933; rcor=3; rNcor=3; rlogoDP=2; rNsEucl=3; rNSW=3; rank_mean=2.800; match_rank=3
a 254 364 432 955 928 613 50 152 0 0 0 0 0 0 0
c 257 170 84 35 40 308 28 247 0 0 0 0 0 0 0
g 262 227 108 1 1 56 884 213 0 0 0 0 0 0 0
t 228 239 376 10 32 24 38 388 0 0 0 0 0 0 0
|
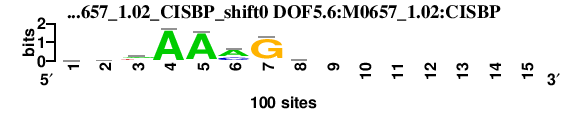
| DOF5.6_M0657_1.02_CISBP_shift0 (DOF5.6:M0657_1.02:CISBP) |
 |
0.768 |
0.410 |
1.691 |
0.908 |
0.932 |
4 |
4 |
1 |
4 |
4 |
3.400 |
4 |
; dyads_test_vs_ctrl_m1 versus DOF5.6_M0657_1.02_CISBP (DOF5.6:M0657_1.02:CISBP); m=4/4; ncol2=8; w=8; offset=0; strand=D; shift=0; score= 3.4; vawAAmGy-------
; cor=0.768; Ncor=0.410; logoDP=1.691; NsEucl=0.908; NSW=0.932; rcor=4; rNcor=4; rlogoDP=1; rNsEucl=4; rNSW=4; rank_mean=3.400; match_rank=4
a 25 36 43 96 93 61 5 15 0 0 0 0 0 0 0
c 26 17 8 3 4 31 3 25 0 0 0 0 0 0 0
g 26 23 11 0 0 6 88 21 0 0 0 0 0 0 0
t 23 24 38 1 3 2 4 39 0 0 0 0 0 0 0
|




